Overview
Our lab studies how cells detect and degrade aberrant RNAs, and how dysregulation of this surveillance process contributes to human muscle development and disease.
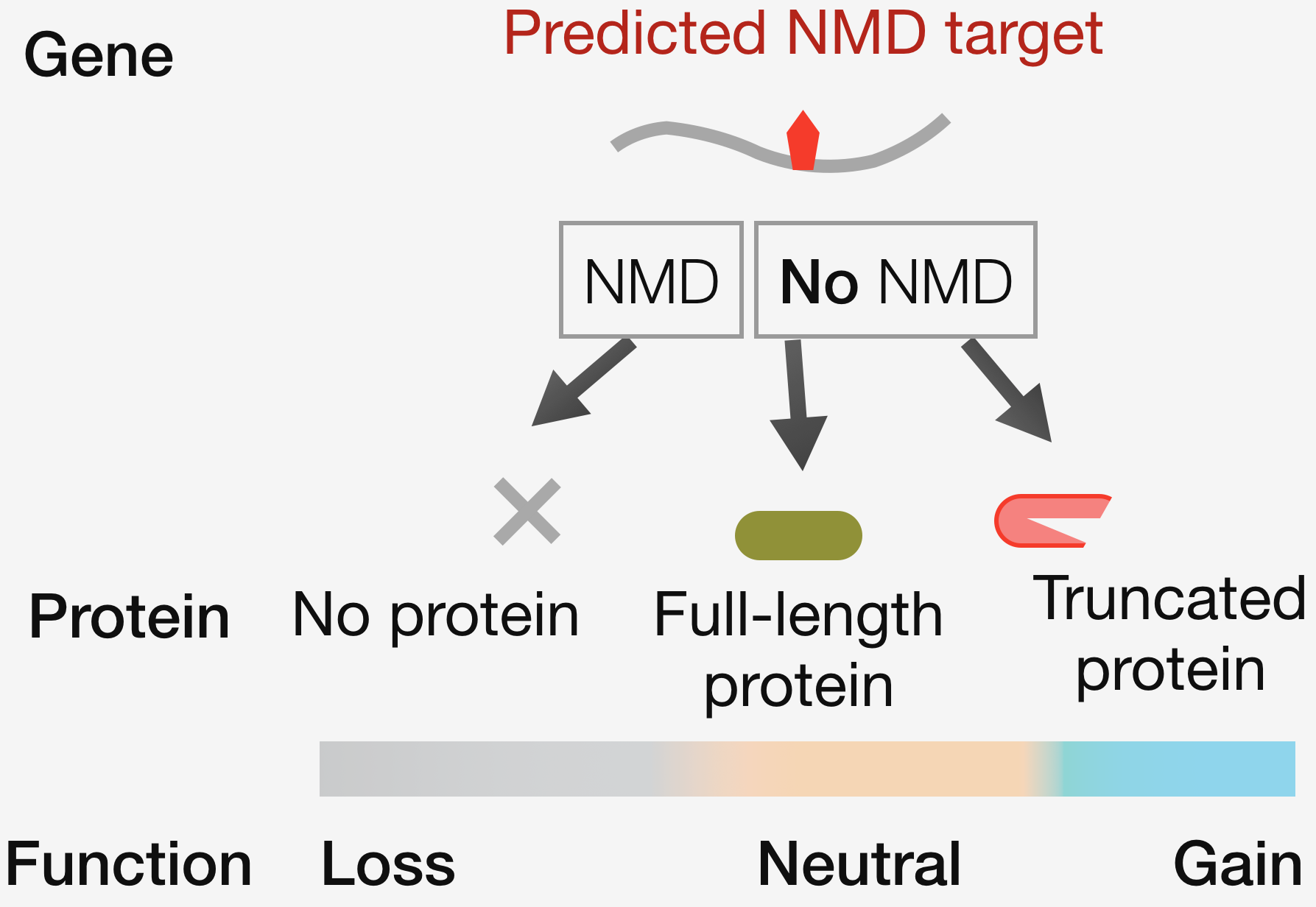
Robustness in biological systems relies on quality control. While errors in the DNA are repaired to maintain fidelity in the flow of genetic information, errors in mRNA are dealt with by degrading the erroneous molecules via surveillance processes such as nonsense-mediated mRNA decay (NMD). Though all mRNAs degraded by NMD follow a set of canonical rules, the converse – that every mRNA that follows the rule is subject to NMD – is not true. This variation in RNA quality control is a major confounding factor in interpreting the clinical impact of human genetic variants. An NMD target may degrade and generate no protein, or escape degradation and result in the production of a full-length or truncated protein – each scenario associated with a potentially different outcome for the cell. The long-term goal of our research program is to understand the molecular logic governing the varying efficiency in RNA quality control and its impact on gene expression, and in turn, cellular phenotypes.
Our questions
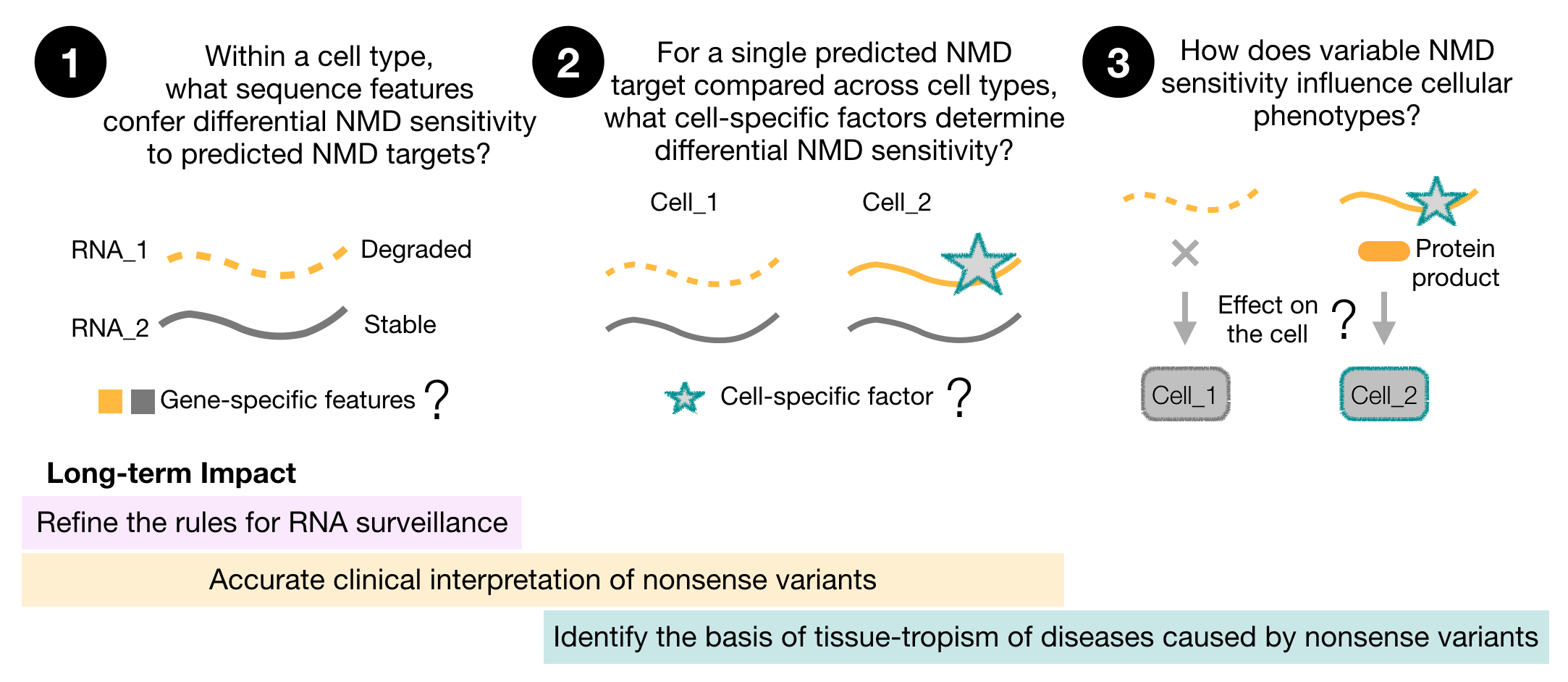
- What cis elements and trans factors allow an mRNA to escape NMD?
- How does variable NMD sensitivity influence cellular phenotypes?
- What is the role of loss of NMD in facioscapulohumeral muscular dystrophy (FSHD)?
Our System
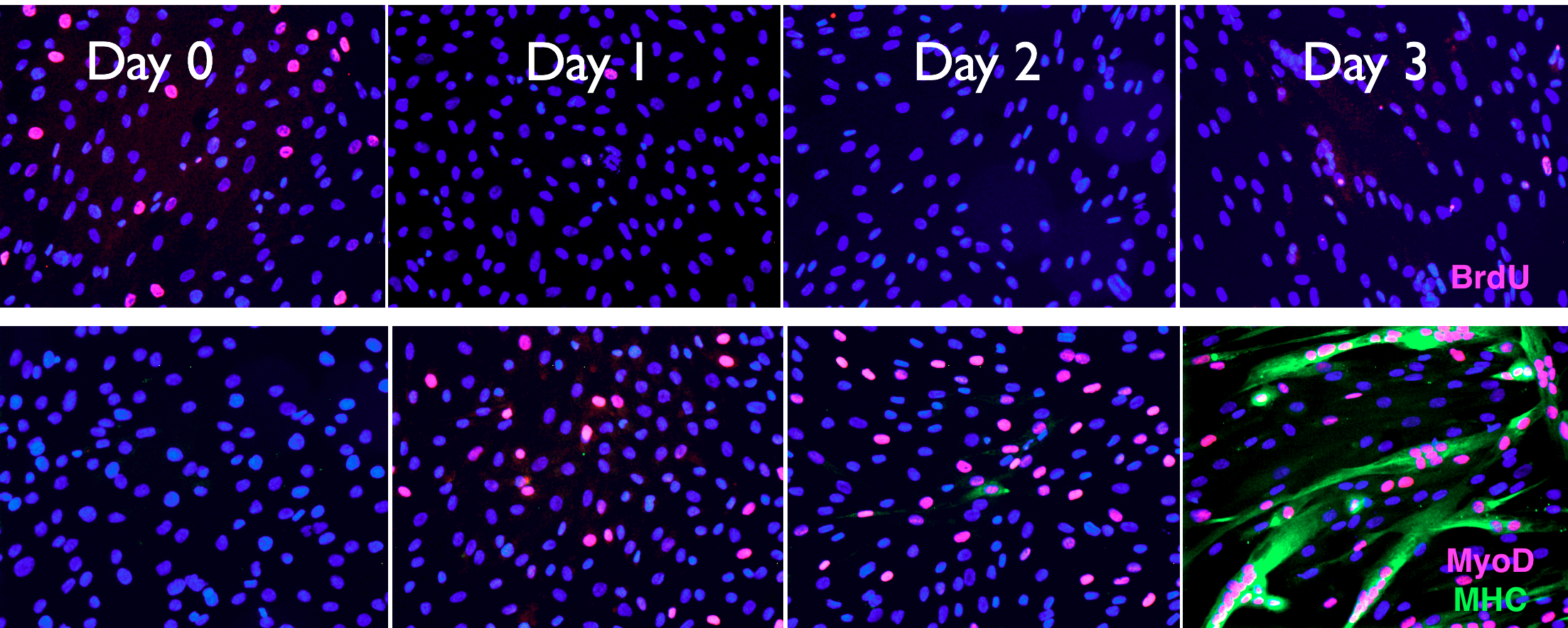
We study RNA surveillance across biological scales - intracellular, tissue-level and across human populations. The system we use the most is the human muscle. Myoblasts that are cultured to confluency fuse and form elongated myotubes upon starvation (image below), enabling us to study both the precursor and differentiated muscle cells in culture under defined conditions.
We utilize both healthy muscle cells and those isolated from individuals affected by FSHD to understand how RNA surveillance contributes to both physiological and pathological processes. During FSHD, the process of NMD is compromised via proteasomal degradation of several key NMD factors. Understanding the underlying mechanism of NMD downregulation by the proteasome, as well as the pathological consequences of NMD inhibition during FSHD could hold essential clues for identifying a cure for FSHD.
Our tools
We apply computational and experimental tools to understand how RNA surveillance shapes gene expression and contributes to disease processes. To this end, we use a wide variety of methods ranging from RNA imaging in single cells to high-throughput genomic analyses.